Annotated libraries

Annotated libraries: a tool for a more informed pharmaceutical screening
This is a short piece on the subject of annotated libraries and the uses they have in drug discovery. It is based on the article “Focused chemistry from annotated libraries” by Balakin et al. Annotated libraries are, in essence, libraries with added data, be it the results of assays, ADME/T or something else of relevance. ChemDiv offers numerous annotated libraries for you to employ in your research.
Classification
Annotated libraries can be divided along the lines of four key characteristics:
-
Size: from a few hundreds to hundreds of thousands molecules
-
Purpose: from extremely specific – target-wise, like for a singular family of kinases, or
characteristic-wise, focusing on the toxicity, for example, to an annotated library with a
broader focus
-
Extent of annotation: from a fully commented library, in which all molecules have all the
fields filled, to a partially commented one, in which some fields in the database will be
empty
-
Data source: from assays to literature
While the classification might feel somewhat dry, it is useful for conveying information effectively, which, in a sense, is the purpose of annotated libraries in general. For example, our human ion channel library contains data on measured activity from different sources for three hundred and twenty molecules, making it a small highly-focused partially covered annotated library with a variety of data sources.
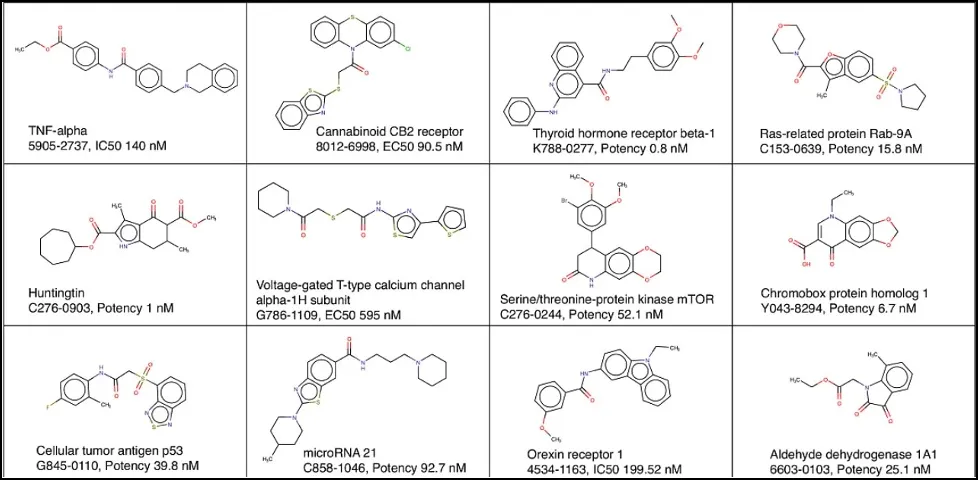
Some examples of molecules in an annotated library, from our ChemoGenomic library, numbering almost ninety-one thousand compounds
Databases built with annotated libraries
Using the aforementioned classification, here we will outline four groups of technology built with annotated libraries:
-
Fully annotated multitoll libraries. For the price of being severely limited when it comes to the choice of compounds, and so only having a relatively small number of them, these annotated libraries provide high quality information with full coverage, making them a good tool for target identification
-
Broad-focus libraries with experimental data. At the cost of inconsistencies, these contain large numbers of entries and give information about actual effects
-
ADME/T annotated libraries. Such libraries do one thing, and do it well – summarize data about pharmacokinetics and help to integrate that information into the workflow. They also may suffer from some internal inconsistencies and only partial annotation
-
Focused annotated libraries. These choose a select number of targets, allowing for a large number of molecules to be included, and have the same problems as the two before.
When paired with the tools of chemoinformatics, annotated libraries become a powerful tool for screening, creation of new libraries or QSAR modeling. From linking gene expression to drug structure to finding the probable mechanism of action or target based on the assay pattern – just to name a few of the applications.
Conclusion
These extremely data-dense annotated libraries help us handle one of the main problems of the current age: instead of the days of old, when the information was hard to come by, we are almost drowning in it. Annotated libraries structure it in a way that allows us to use all the information available to the full extent and to connect all sources and researches together. Even though this approach is not without downsides, namely the internal inconsistencies within libraries, which complicate further analysis, they are a potent tool for drug discovery. Aside from the two examples given here, ChemDiv offers many more annotated libraries – from a CNS library to one focused on the opportunistic pathogens.


